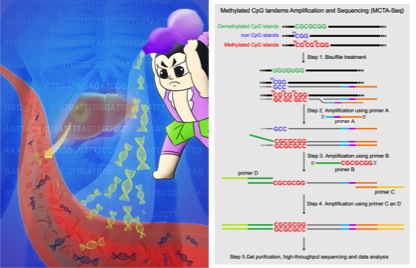
Analyzing DNA methylation in circulating cell-free DNA (ccfDNA) is hampered by the low throughput of current methods. Now reported in the November 2015 Issue of Cell Research, a collaborative team of Biodynamic Optical Imaging Center (BIOPIC) of Peking University and Beijing Shijitan Hospital of Capital Medical University (the Ninth Clinical Medical School of Peking University) developed a Methylated CpG Tandems Amplification and Sequencing (MCTA-Seq) method to enable genome-scale detection of hypermethylated CpG islands in ccfDNA. Application of this method for hepatocellular carcinoma (HCC) achieved noninvasive early detection of the cancer.
DNA methylation is the epigenetic modification of the cytosine to the 5’-methylated-cytosine by adding a methyl group and mainly occurs at the CpG dinucleotide. The CpG dinucleotide accumulates at the genomic region called CpG islands, which are generally located at the gene promoter and essential for transcription regulation. CpG islands are usually not methylated in the normal cell, while in cancer cells, a large number of CpG islands gain methylation. The CpG islands hypermethylation not only plays important roles in cancer development, but also is a promising cancer biomarker.
Dying cancer cells will shed their DNA to blood as ccfDNA. Is it possible to detect cancer by analyzing the ccfDNA carrying the hypermethylation information? Although researchers have been striving for this since the last twenty years, the progression is slow with one key bottleneck being a lack of a genomic method. The ccfDNA released by an early cancer is scanty and highly fragmented, which goes beyond the detection sensitivity of current DNA methylome technologies.
MCTA-Seq overcomes the bottleneck using a clever strategy. Through selectively amplification of the methylated CpG tandems CGCGCGG sequence, it can detect nearly 9,000 hypermethylated CpG islands in one reaction with the low detection limits being as little as the genomic DNA of one to two cells.
The researchers applied MCTA-Seq to hepatocellular carcinoma (HCC), which is one of the most common malignant tumors worldwide. Due to the prevalence of chronic HBV infection, the occurrence is particularly high in East Asia including China. The serum α-fetoprotein (AFP) is the most commonly used biomarker for detecting HCC; however, it has false negative results in 40% HCC patients. Therefore, it is urgent to develop novel HCC biomarkers.
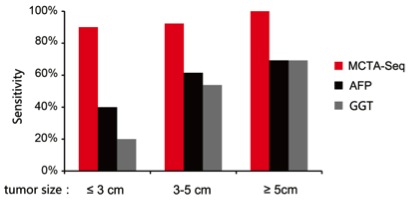
The researchers analyzed a total of 151 clinical samples including 57 tissue samples, and 94 plasma samples obtained from HCC patients, cirrhosis patients and healthy individuals. In the HCC tissues, they identified nearly 900 aberrantly hypermethylated CpG islands. From the plasma samples, they found that nearly 400 CpG islands are differentially hypermethylated in small HCCs (≤ 3 cm); by raising the criteria, they got 41 top HCC-detecting markers. Interestingly, they found that the plasma methylation markers can be divided into two types. One type directly comes from the cancer tissue, while the other type is derived from both the cancer tissue and the non-cancerous liver tissue. In plasma of some small HCC patients, the later one elevates at a level more prominently than the former one. Both types of markers drop after resection of the cancer, indicating that they are tightly associated with the cancer. The latter group of HCC markers, which has not been described previously, shows the power of MCTA-Seq as a screening method without preconception.
By combining two types of markers, MCTA-Seq detects HCC with a sensitivity of 94% at a specificity of 89%. Importantly, MCTA-Seq successfully identified all fifteen AFP-negative patients.
Since CpG island hypermethylation occurs in most human cancer types, MCTA-Seq also offers promising for detecting other cancer types in blood. Further, since DNA methylation has both tissue and tumor-specific patterns, the rich information provided by MCTA-Seq give the potential to recognize the tissue origin of a cancer in blood. MCTA-Seq is also simple and cost-effective with a single-tube reaction and a relatively low sequencing depth.
Dr. Lu Wen and graduate student Jingyi Li, Xiaoming Liu and Huahu Guo are co-first authors of this paper. Prof. Fuchou Tang and Prof. Yanyi Huang at BIOPIC of Peking University and Prof. Jirun Peng at Beijing Shijitan Hospital of Capital Medical University are co-correspondence authors. This work is supported by NSF China.
Lu Wen*, Jingyi Li*, Huahu Guo*, Xiaomeng Liu*, Shengmin Zheng, Dafang Zhang, Weihua Zhu, Jianhui Qu, Limin Guo, Dexiao Du, Xiao Jin, Yuhao Zhang, Yun Gao, Jie Shen, Hao Ge, Fuchou Tang#, Yanyi Huang#, and Jirun Peng#. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Research. (2015) 25: 1250–1264. doi:10.1038/cr.2015.126; published online 30 October 2015.