How a cell can become plastic and change their identity has fascinated scientists for decades. In vivo lineage reprogramming, conversion of a highly specialized cell into the desired cell identity in a living body, has recently emerged as a promising regenerative strategy. However, molecular mechanisms underlying in vivo lineage conversion remained unclear. Natural lineage reprogramming, the conversion of one cell identity to another during normal development, although rare, occurs with high efficiency and temporospatial precision. These natural lineage conversion events therefore provide powerful model systems for deciphering the mystery of cellular plasticity.
Temporospatial induction of homeodomain gene cut dictates natural lineage reprogramming
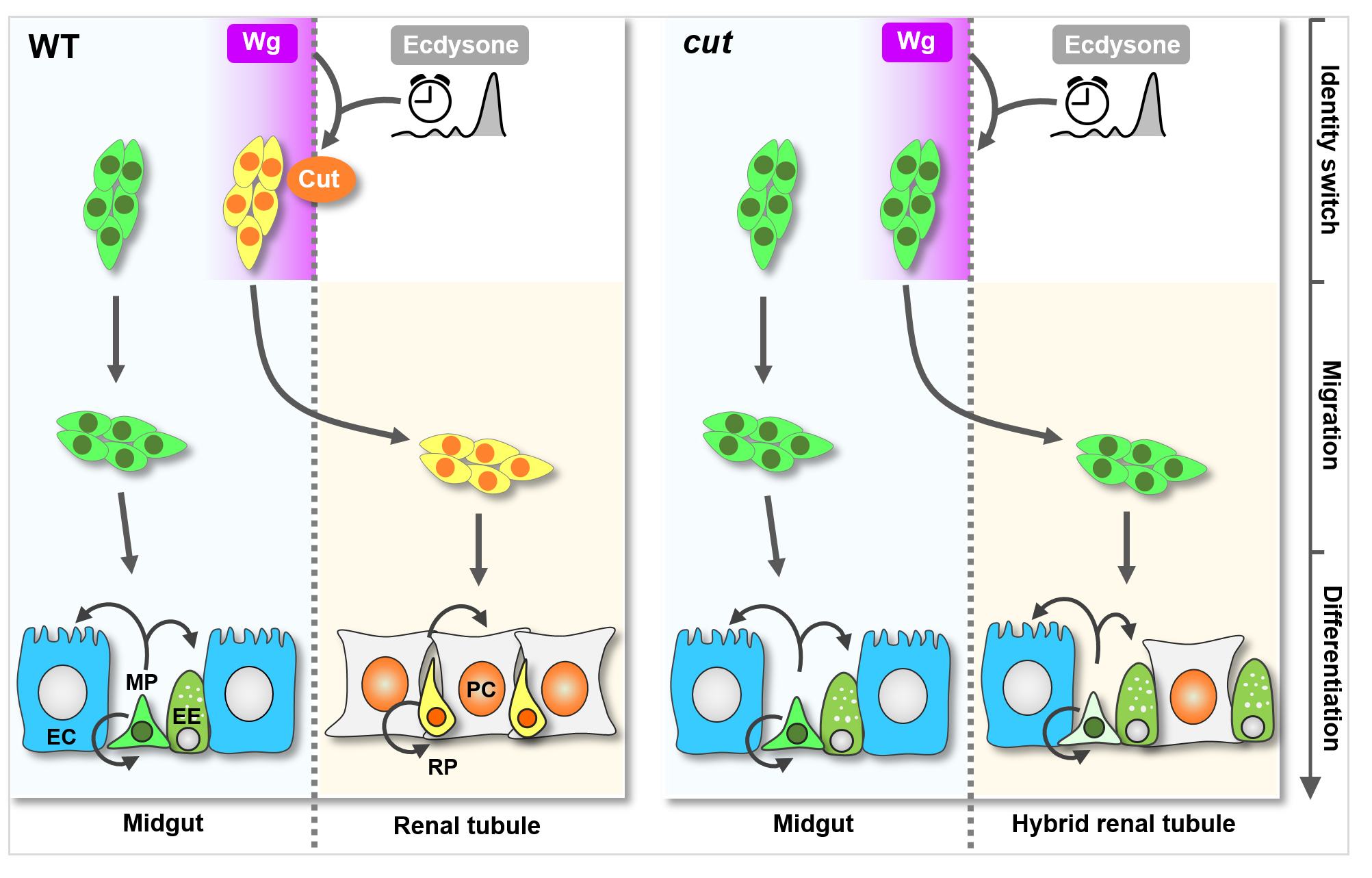
Dr. Yan Song’s group at the School of Life Sciences and Peking-Tsinghua Center for Life Sciences has recently published a Research Article at eLife titled “Temporospatial induction of homeodomain gene cut dictates natural lineage reprogramming”. This work uncovered a natural midgut-to-renal lineage reprogramming event during fruit fly metamorphosis and identified a highly conserved protein called Cut as a crucial cell identity switch in this process. A steep gradient of a spatial molecule (called Wingless/Wg) intersects with a pulse of steroid hormone to induce Cut expression specifically in a small subset of fly midgut progenitors, precursors that produce functional midgut cells, and convert them into renal identity. These reprogrammed progenitors in turn migrate across organ boundary and onto renal tubules, where they give rise to functional renal cells. Strikingly, midgut progenitors depleted of Cut migrate normally onto renal tubules, yet fail to switch their cell identity and give rise to midgut cells along renal tubules, causing renal identity crisis.
Intriguingly, this work found that the temporal and spatial signals triggering this natural lineage reprogramming event likely intersect through an unexpected chromatin looping mechanism. That is, the temporal factor not only form protein complex with the spatial factor, but also bridges two important yet far-away DNA regulatory sequences, called promoter and enhancer, of cut gene together via self-association, inducing cut gene activation. This mechanism might represent a new and general paradigm governing cell fate or identity switch with high precision in space and time, and has promising implications for the development of regenerative medicine strategies.
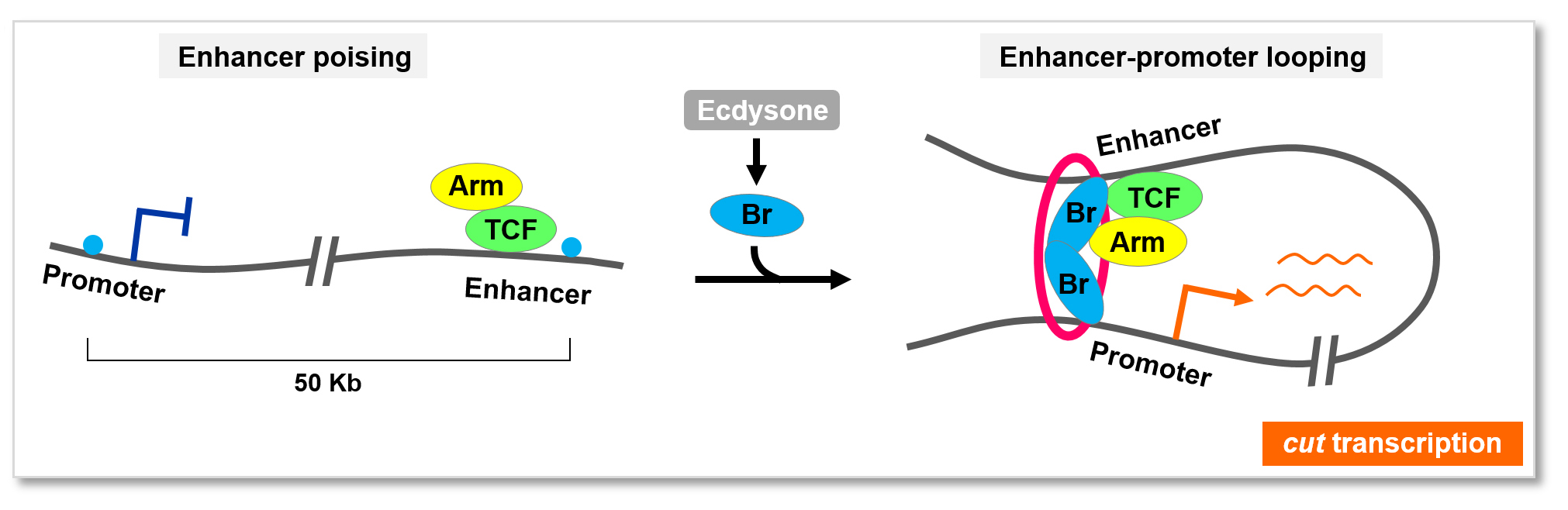
A “poising and bridging” model
The work was published online in eLife with PhD Student Ke Xu as the first author and Yan Song as the corresponding author. The work was supported by grants from National Natural Science Foundation of China, Peking-Tsinghua Center for Life Sciences and the Ministry of Education Key Laboratory of Cell Proliferation and Differentiation.